 © Isabelle Domaizon
© Isabelle Domaizon
Quanti-Inventory
Consolidating quantitative methods for monitoring fish communities using eDNA: validation of methodological choices
SECTIONInnovative monitoring |
PERIODJanuary 2024 - January 2025 |
|
FUNDINGPôle ECLA |
STUDY AREA(S)Hexagonal France |
|
PARTNERSUniversité Savoie Mont Blanc, INRAE, UMR CARRTEL, OLA 74200 Thonon-les-Bains, FranceService EcoAqua, DRAS, OFB, Aix-en-Provence, France Aix Marseille Univ., INRAE, RECOVER, 13 182 Aix-en-Provence, France Jean Guillard INRAE, UR-Riverly, Centre de Lyon-Villeurbanne, Villeurbanne Cedex, France |
COORDINATOR(S)I. Domaizon |
Project Description
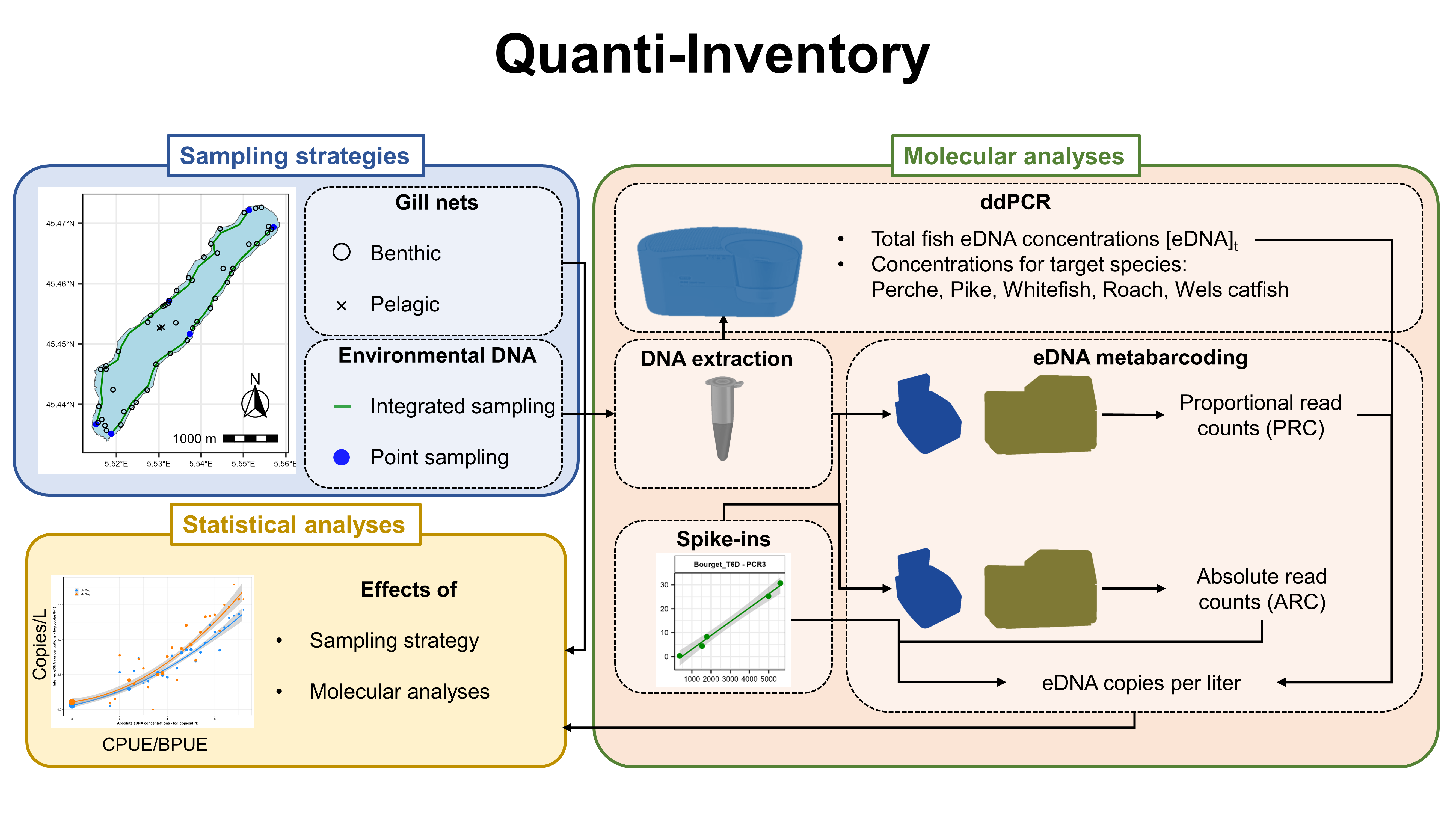
Project Context :Attempts have been made to obtain quantitative data from eDNA metabarcoding surveys for fish communities. These approaches make it possible to convert DNA read counts into DNA copy numbers for each species and are a better approximation of the abundance-biomass of the species. One method combines metabarcoding data with quantitative PCR analyses (Pont et al 2022), while the other method involves adding internal standards (known quantities of DNA from ‘exotic’ species) to each sample before sequencing (Ushio et al 2018). A formal comparison and validation of these two approaches is currently lacking.
Project Objectives :The aim of the Quanti-Inventory project is to carry out a formal comparison in order to develop a more quantitative eDNA biomonitoring tool. The ultimate goal is to be able to make informed recommendations for an operational protocol that allows for more quantitative monitoring of lake fish communities. In the short term, this protocol, used in combination with other non-invasive techniques such as hydroacoustics, should provide a relevant tool for biomonitoring fish communities in lakes.
Project Results
To go further
Useful Links :
https://doi.org/10.1101/2025.09.05.674411
Contact : isabelle.domaizon[at]inrae.fr